Stelios Ninidakis, HCMR
This experiment generated results on marine biodiversity at different stations across the European coastline. The input data come from EMBRC’s project, the European Marine Omics Biodiversity Observation Network (EMO BON: https://www.embrc.eu/services/emo-bon). Assessing marine biodiversity data connects to effective protection of natural biodiversity and sustainable management of marine ecosystems. In addition, coastal tourism and industries, such as fishing, aquaculture, and pharmaceuticals, depend on healthy and diverse marine ecosystems. This work uses reliable and frequent data to provide robust data products aiming to increase knowledge on marine biodiversity and deepen understanding of species’ interaction and connection to stressors. The results of this experiment will feed several EU Horizon projects for the development of Virtual Research Environments (VREs). In addition, results will become available to a wide community of scientists and experts to work further on them. We envision running events (datathons) with experts from different fields to work on the data by topic of interest and to respond to pending scientific questions. This experiment would be unfeasible to run on a standard personal computer, as it needs more than 6 days running on a 40-core Intel(R) Xeon(R) Gold 6230 CPU @ 2.10GHz in analyzing the dataset that was used to derive the first data products. Special thanks to MeluXina supercomputer (https://docs.lxp.lu/) for providing access to the CPU partition.
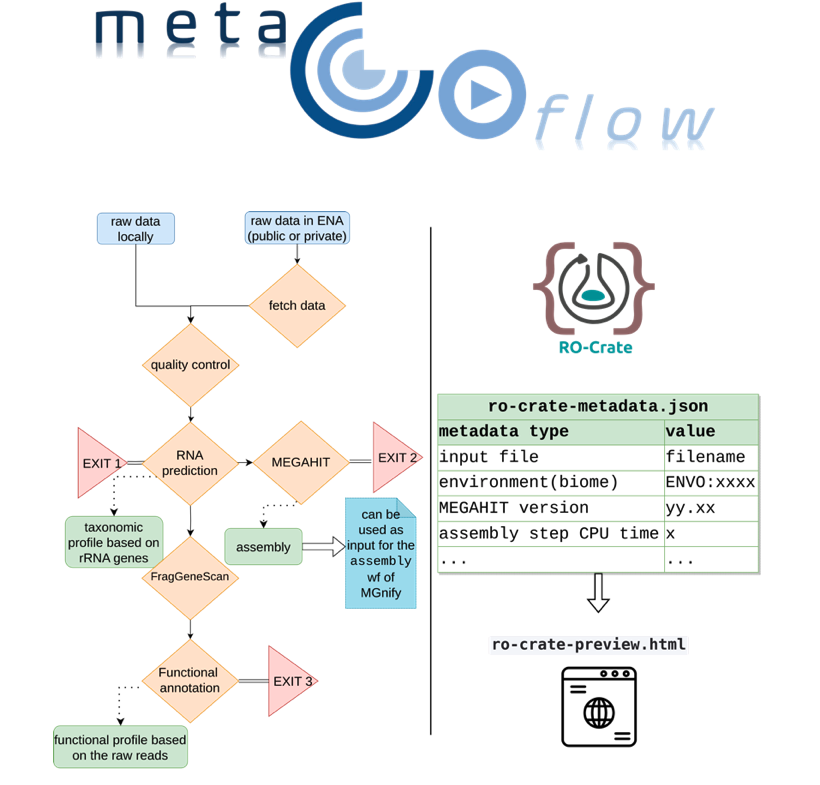
Haris Zafeiropoulos, Martin Beracochea, Stelios Ninidakis, Katrina Exter, Antonis Potirakis, Gianluca De Moro, Lorna Richardson, Erwan Corre, João Machado, Evangelos Pafilis, Georgios Kotoulas, Ioulia Santi, Robert D Finn, Cymon J Cox, Christina Pavloudi, 2023. metaGOflow: a workflow for the analysis of marine Genomic Observatories shotgun metagenomics data. GigaScience, 12, p.giad078. Available on: https://academic.oup.com/gigascience/article/doi/10.1093/gigascience/giad078/7321054