High-Performance Computing Enhances Treatment Precision in Breast Cancer
Presentation of the problem and objective of the experiment

In cancer treatment, it is a clinical challenge to select the right drug which will work for a specific patient, as the efficacy of cancer drugs is highly dependent on individual characteristics on a molecular level. The solution developed by the experiment helps to narrow down the most promising drugs, using an HPC-backed machine learning approach to analyse a huge initial dataset. The aim is to implement an easy-to-use and intelligent platform which can identify the drugs most likely to achieve high effectiveness in each individual patient based on the specific patient’s molecular profiles.
Organisations involved
Short description of the experiment
Breast cancer continues to be a major health issue globally and many cancer patients fail to respond to their treatment. This lack of efficacy is mainly attributed to host/tumour variations at the genetic and molecular level, which clinical practice still struggles to address. The emergence of new genomic technology combined with digitalization has delivered treatment regimens that assess the DNA, RNA, protein, and metabolites in the individual patient’s tumor and integrate those into therapeutic decision-making. However, current technologies focusing on just one or a few genetic biomarkers or using complex ex vivo laboratory tumor models are predictive of treatment outcomes only in highly selected cases and difficult to implement effectively.
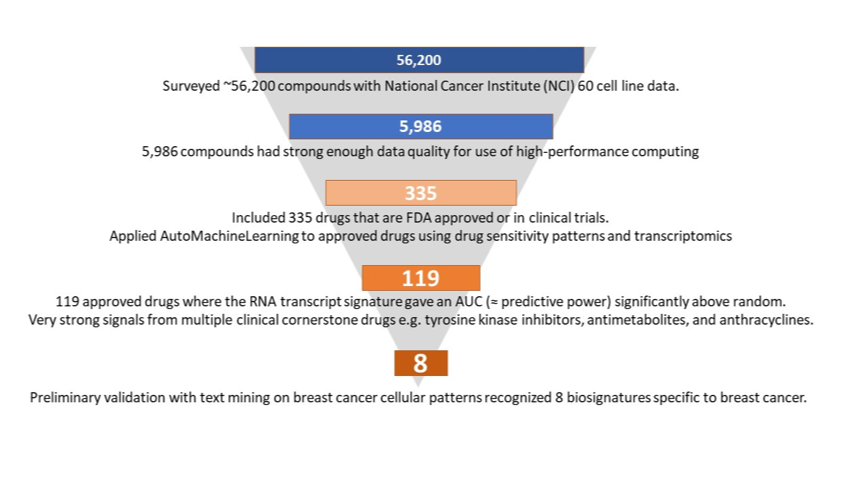
The experiment carried out extensive analyses of a huge volume of publicly available data. The NCI-60 data set links 60 human cancer cell lines representing different types of cancer to the anticancer activity of over 50,000 compounds. Using specific quality criteria, which were defined at the start of the experiment, 5,986 compounds out of those over 50,000 were selected for further analysis, including 335 drugs that are FDA-approved or in trials. Using the JADBio autoML platform and HPC resources, ML models for the selected compounds were built to estimate the models’ performance in predicting treatment outcomes. As a means of early validation of the ML models, biological text mining was carried out independently. It revealed eight specific models which are particularly interesting for breast cancer, which was among the promising 119 models also identified by ML. hey include models for key anticancer drug classes used in breast cancer, corroborating the value of the HPC-backed ML approach and building the basis for further clinical validation.
Outlook
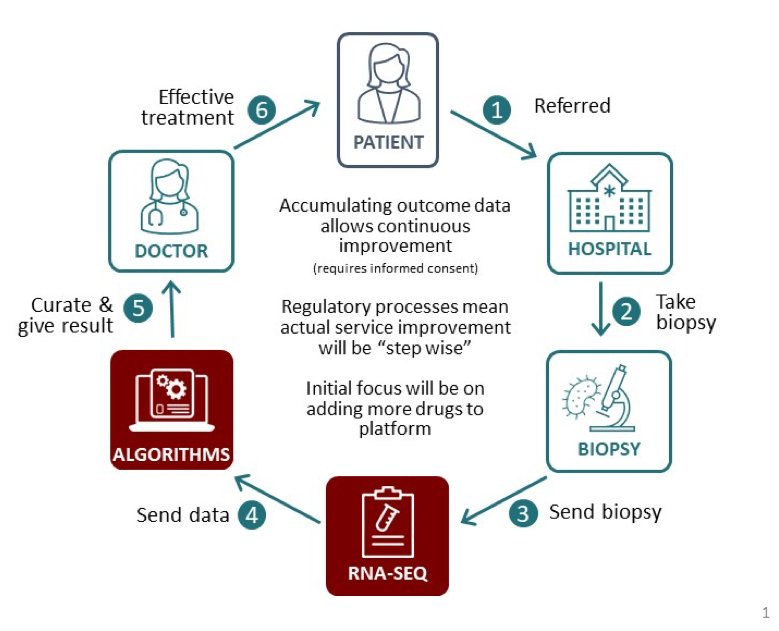
After further validation, the models will be used to set up a complete platform called ‘Allied Intelligence for Drug Accuracy’ (AÏDA) which predicts the efficacy of different cancer drugs for each individual patient, based on their biopsy readings. Clinicians will receive a report listing a large number of relevant drugs that highlight those most likely to work for a given patient’s cancer.
Lessons learned
The experiment carried out extensive analyses of a huge volume of publicly available data (called NCI-60) which would have required a prohibitive amount of time without the employment of HPC.
Expected impact
Social
The AÏDA technology has a huge potential to support clinicians in their choice of treatment. No similar solutions exist at the moment and therefore AÏDA has the opportunity to become a first-in-market product that can truly revolutionize the way cancer patients are treated, providing clinicians with an unrivalled recommendation service for cancer drugs, based on individual patient’s biopsy.
Business
With a breast cancer incidence of over 780,000 in 2018 in the EU and USA alone, there is a huge market potential to be exploited with such a commercial response prediction test – even using very conservative assumptions. The market launch is expected in mid-2024 in Germany and Nordic countries, where 23,000 cases of breast cancer are newly diagnosed per year, offering a business potential of up to €69m, based on an anticipated price of €3,000 per service.
Exploitation roadmap
The business model is highly scalable and the system can be applied to any tumor type and any drug that has demonstrated toxicity. Besides direct economic and clinical benefits, all partners will enjoy increased visibility in the biomedical market and scientific community, generate new intellectual property, and foster company growth. The HPC-based solution can play a role as a use case for promoting other diagnostic/prognostic/predictive applications in the field of personalized medicine, fostering wider application.
HPC enables and accelerates data-driven solutions in biomarker discovery towards precision medicine. – Katerina Hatzaki, DUTH








